
Drug Target Models
GPCR Reporter Cells
Immunotherapy cells
Other Stable Cells
Assay Kits & Reagents
Services
Resources
Company
Professional technology integration to support the entire R&D process
Transposon-Based Integration
Product Description
Cycle Pricing
Case Studies
Definition
Transposable elements (TEs), also known as jumping genes, are DNA sequences capable of moving from one genomic location to another. First discovered by Barbara McClintock, TEs are now recognized as ubiquitous components of genomes across both prokaryotes and eukaryotes. In many organisms, they constitute a substantial proportion of the genome—for example, approximately 50% of the human genome and up to 90% of the maize genome.
Transposon Types
We often use the transfection of chemical reagents have Lipofectamine/CaPO4 / FuGene/PEI, basic principle is the same:
Transposable elements can be divided into two categories based on their transposition mechanism:
1. Class I transposable elements (also known as retrotransposons)
2. Class II transposable elements (also known as DNA transposons). 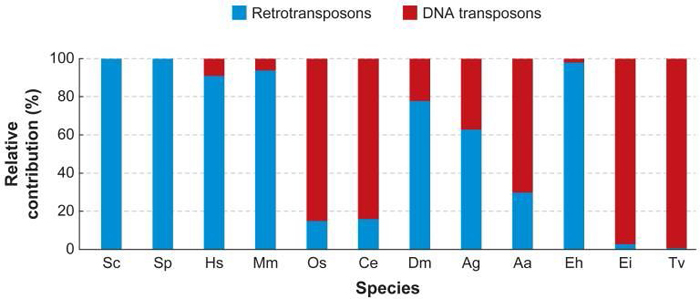
1. Class I transposable elements (also known as retrotransposons)
2. Class II transposable elements (also known as DNA transposons).

Figure 1: Relative abundance of retrotransposons and DNA transposons in the genomes of different eukaryotic organisms.
This figure shows the percentage of DNA and retrotransposons relative to the total number of transposable elements in each species. (Sc: Saccharomyces cerevisiae; Sp: Schizosaccharomyces pombe; Hs: Homo sapiens; Mm: Mus musculus; Os: Oryza sativa; Ce: Caenorhabditis elegans; Dm: Drosophila melanogaster; Ag: Anopheles gambiae (malaria mosquito); Aa: Aedes aegypti (yellow fever mosquito); Eh: Entamoeba histolytica; Ei: Entamoeba invasives; Tv: Trichomonas vaginalis.)
Class I Transposable Elements (TEs): Retrotransposons
Class I transposons, also known as retrotransposons, transpose via a "copy-and-paste Ctrl C+V" mechanism (Figure 1). They first replicate themselves as RNA transcripts, which are then reverse-transcribed back into DNA by an enzyme called reverse transcriptase, ultimately inserting into a new target site. This is similar to the replication mechanism of retroviruses such as HIV.
Class I transposons do not encode a transposase. Class I TEs are considered replicative because they replicate with each jump. This increases the TE copy number and, consequently, the size of the host genome.
Class I transposons come in two types: those with long terminal repeats (LTRs) and those without (non-LTR transposons). LTR retrotransposons have a structure and replication mechanism similar to retroviruses. They contain two genes: gag and pol. The pol polyprotein encodes the reverse transcriptase and integrase required for transposition in LTR retrotransposons. Non-LTR retrotransposons contain two open reading frames (ORFs), typically ending in poly(A). ORF2 encodes endonuclease and reverse transcriptase activities.
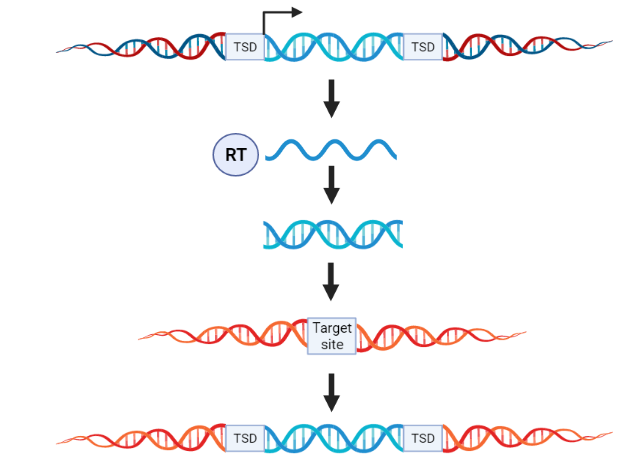
Figure 2: Overview of retrotransposon transposition.
Retrotransposons are moved using a “copy and paste Ctrl C+V” mechanism.
Class II TEs: DNA transposons
Class II transposons are also called DNA transposons because they do not require RNA mediation for their movement. Most class II transposons employ a non-replicative "cut and paste Ctrl X+V" transposition mechanism: they excise themselves from one location and then insert themselves at another (Figure 2). Class II transposons have long terminal repeats (LTRs) at both ends.
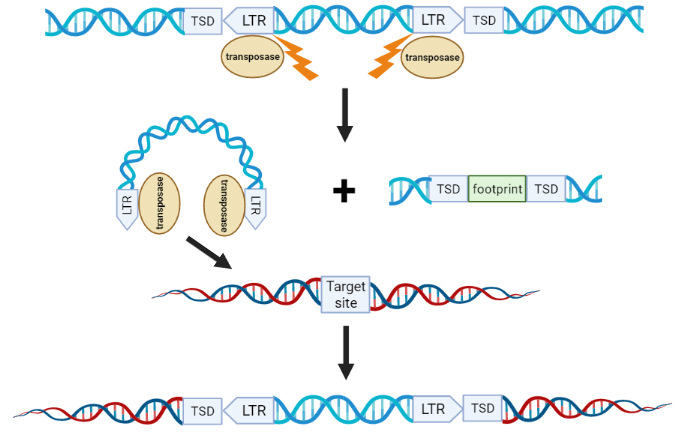
Figure 3: Overview of DNA transposon transposition.
To mobilize a transposon, a transposase first binds to the transposon's long terminal repeat (LTR), inducing a double-strand break (DSB) and excising the transposon from the donor DNA. This leaves behind a DNA footprint. When the transposon-transposase complex finds its target site, it integrates, creating a target site duplication (TSD).
DNA transposons commonly used in the laboratory
Although there are many different types of transposons, DNA transposons are most commonly used for genome manipulation in the laboratory. When using transposons in the laboratory, the transposase gene is provided in trans to insert the target gene between the transposon's long terminal repeats (LTRs), similar to the packaging process used in viral vectors.
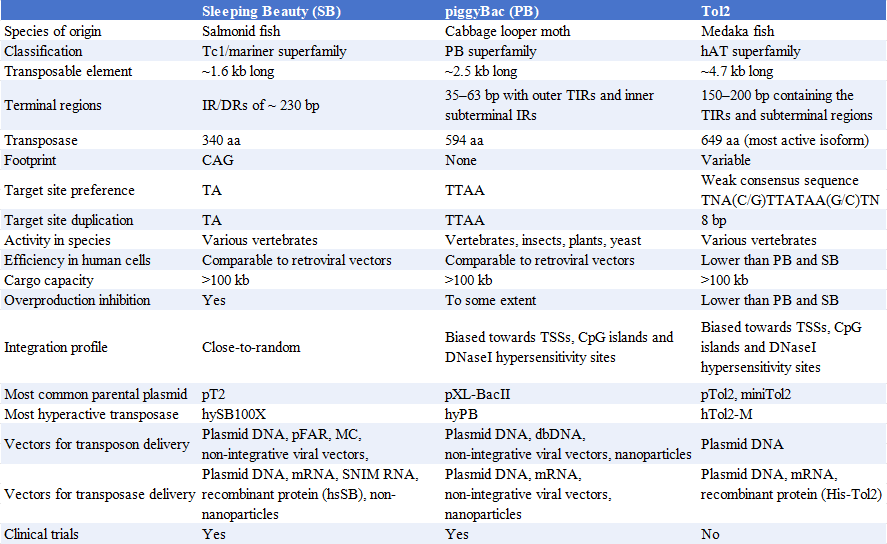
Three common transposon systems suitable for use as research tools are Sleeping Beauty, PiggyBac, and Tol2.
1. Sleeping Beauty (SB)
Sleeping Beauty is a synthetic transposable element derived from mariner (S. marina). Its preferred integration target is the TA dinucleotide. After cleavage by the transposase, it leaves a CAG DNA footprint at the end of the excision site. Its cargo capacity is >100 kB, but integration efficiency decreases with cargo size. Sleeping Beauty integrates into mammalian genomes with near-random integration characteristics. SB is active in vertebrates and integrates in human cells at a rate similar to that of retroviral vectors. The highly active version of SB transposase, SB100X, boasts approximately 100-fold higher efficiency than the first-generation SB transposase. hySB100x is an improvement on SB100X, boasting a 30% increase in transposition activity.
2. piggyBac
PiggyBac, discovered in Pieris rapae, targets the TTAA site. Unlike other transposons, it leaves no DNA footprint after excision. piggyBac can load DNA exceeding 100 kB and is active in yeast, plant, insect, and mammalian cells (including human) both in vitro and in vivo. piggyBac prefers integration at transcription start sites, CpG islands, and DNase I hypersensitive sites. Like Sleeping Beauty, piggyBac integrates in human cells with similar efficiency to retroviral vectors. Compared to the codon-optimized wild-type piggyBac transposase, the highly active PB transposase (hyPB) exhibits approximately 10-fold higher activity in mammalian cells.
3. Tol2
Tol2 was the first active DNA transposon reported in vertebrates. It was discovered in the Japanese medaka fish, where insertion into the tyrosinase gene caused albinism. Unlike Sleeping Beauty and piggyBac, Tol2's preferred integration site, TNA(C/G)TTATAA(G/C)TNA, has a weaker consensus sequence. Tol2 can deliver 10-11 kB of DNA into mammalian cells without loss of efficiency, with a maximum load of approximately 200 kB. Similar to piggyBac, Tol2 also prefers integration at transcription start sites, CpG islands, and DNase I hypersensitive sites. Tol2 is active only in vertebrates, and its integration efficiency in human cells is lower than that of piggyBac and Sleeping Beauty. Minimal Tol2, or miniTol2, is a truncated version of the original Tol2 with approximately three-fold increased transposition activity.
Common Applications
Transposon Mutagenesis Screens
Transposons are inherently mutagenic, making them excellent tools for mutagenesis screens to detect loss-of-function or gain-of-function mutations. In these screens, transposons encode reporter genes, mutagenic cassettes, or barcodes. When delivered to cells or model organisms, they integrate into the host genome. Next-generation sequencing is then used to detect transposon insertion sites, and analysis is performed to determine which insertions were positively or negatively selected during the experiment.
Transgenic Animals
Transgenic animals are typically generated by injecting DNA directly into the pronucleus of fertilized eggs, resulting in random integration of the sequence into the genome, a highly unpredictable process. However, transposons can efficiently integrate into the genome of fertilized eggs after injection into the cytoplasm, a process that is much less efficient when DNA is injected directly. Sleeping Beauty, piggyBac, and Tol2 have all been used to generate transgenic animals, including zebrafish, mice, rats, and rabbits.
Stable Cell Line Construction
Transposons are an alternative to lentiviral vectors for stable cell line construction, such as for iPSC reprogramming and gene and cell therapy, and they have the potential to overcome some of the limitations of viruses. Transposons (TEs) have large payloads, reaching up to 100 kB using Sleeping Beauty and piggyBac, offering a significant advantage over viral vectors (AAV payloads are approximately 5 kB, while lentiviral payloads are approximately 8 kB). Transposons are also less likely to induce an immune response than viral vectors and are easier and cheaper to produce. Both delivery methods can potentially cause gene disruption due to integration, but because TEs primarily insert into intergenic regions, gene disruption is less of a concern
sgRNA-guided Site-Specific Transposition
SgRNAs can be used to guide transposition at a specific location. The INTEGRATE system (guide RNA-assisted targeted insertion of transposable elements) has been reported to achieve approximately 100% integration of DNA fragments up to 10 kB in bacteria.
Stable Cell Line Construction Services
Reqbio provides stable cell line development services based on the "Transposon Integration System." To date, we have successfully generated over 300 stable cell line models, demonstrating extensive experience in electroporation-based genome integration. Please contact us to discuss your project requirements.
Definition
1、Random integration has low integration efficiency and may not necessarily integrate into the transcriptional active regions.
2、Random integration has low integration efficiency and may not necessarily integrate into the transcriptional active regions.
Definition
1、Random integration has low integration efficiency and may not necessarily integrate into the transcriptional active regions.
2、Random integration has low integration efficiency and may not necessarily integrate into the transcriptional active regions.
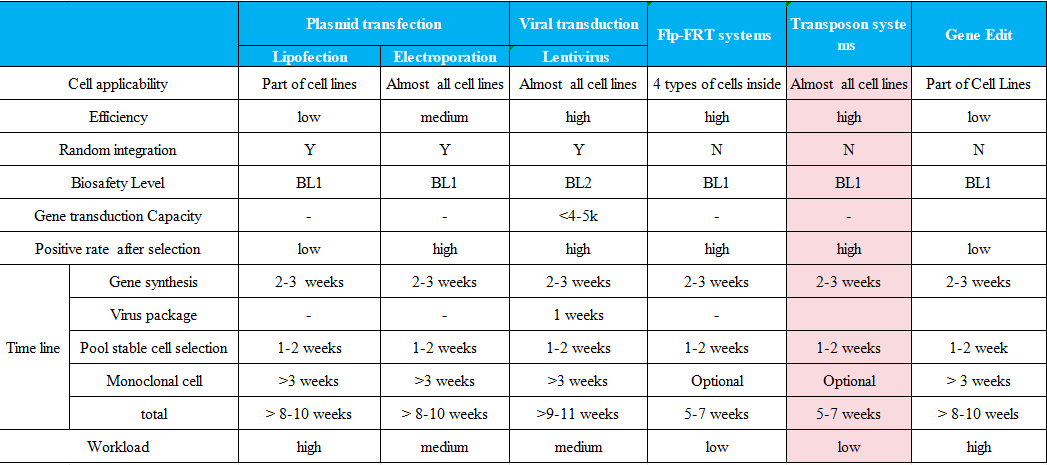
Other stable cell lines
Lentiviral Integration
Plasmid Electroporation
Flp-FRT Targeted Integration
Random Plasmid Integration
If you are interested in ordering, please contact us.
Customer help-line
4008-750-250
sales@reqbio.com
Office address:
3rd Floor, No. 6, Lane 222, Guangdan Road, Pudong New Area, Shanghai, China
We Are Pleased to Announce: Global Commercial Licensing Rights for Jurkat E6.1, CHO-K1, and HEK293 Cell Lines Officially Secured.
Explore