
Drug Target Models
GPCR Reporter Cells
Immunotherapy cells
Other Stable Cells
Assay Kits & Reagents
Services
Resources
Company
Professional technology integration to support the entire R&D process
Digital PCR (dPCR)
Product Description
Cycle Pricing
Case Studies
Service Principle
Droplet Digital PCR (ddPCR) is a digital PCR method developed by Bio-Rad that utilizes a water–oil emulsion droplet system. In this system, the sample is partitioned into thousands of nanoliter-sized droplets, each serving as an individual reaction chamber, similar to a test tube or well in a PCR plate but on a much smaller scale. This extensive sample partitioning is a key feature of ddPCR, enabling precise and absolute quantification of target DNA molecules.
DdPCR Advantage
Absolute quantification
ddPCR provides absolute counts of target DNA per sample without the need for a standard curve, making it an ideal technology for measuring target DNA, analyzing viral loads, and quantifying microbial populations.
High precision
The large number of sample assignments provided by ddPCR can reliably measure small differences in the copy number of the target DNA sequence in the sample.
Improved signal-to-noise ratio
High-copy templates and background DNA are diluted, enhancing the concentration of template molecules in target-positive partitions. This enables sensitive detection of rare targets with quantitative accuracy of ±10%, a sensitivity down to a single nucleic acid molecule, and a detection limit as low as 0.001%.
Eliminate PCR bias
Eliminate qPCR amplification efficiency dependence, reduce the error rate, and detect small (1.2 times) differences, here it should be pointed out.
Reduced consumable costs
Reaction volumes in the picoliter to nanoliter range reduce reagent usage and sample volume per data point, especially for precious samples.
Excellent partitioning
ddPCR generates approximately 20,000 droplets per 20 µl sample, allowing nearly 2 million partitioned PCR reactions on a 96-well plate. Greater partitioning directly enhances quantification precision.
Service Application
- Detection of Gene Expression, mainly quantitative analysis of mRNA detection.
- Detect Mutation (SNV and Indel), can detect copy number, and can also detect ratio.
- Detect Fusion, can detect the copy number, and can also detect the ratio.
- Detection of CNV (copy number variation analysis), relative quantification.
Experiment 8:Reproducibility test
Take 10 standard samples with different AF (1%~50%) and arrange ① Operator A and Operator B to perform four experimental tests each on the same day; ② Operator A performs four experimental tests on the first day and the second day respectively; ③ Operator A performs four experimental tests on the first day and Operator B performs four experimental tests on the second day.
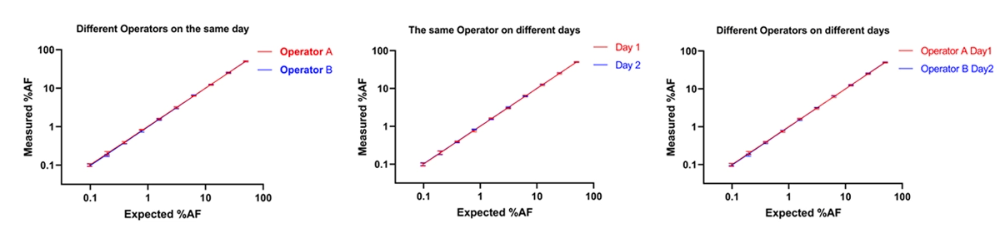
From the above data, it can be seen that the EGFR T790M detection system is stable and reproducible in different people and at different times.
Experiment 7:Repeatability test
Take 10 standard samples with different AF (1%~50%) and arrange Operator A and Operator B to perform four tests on the same day.
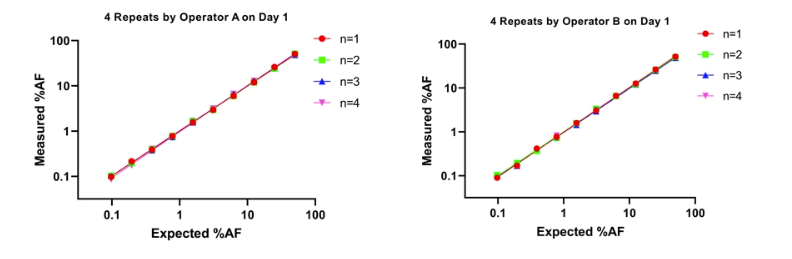
It can be seen from the above data that the EGFR T790M detection system is stable and reproducible.
Experiment 6:Recovery test
First, the corresponding negative standard of EGFR T790M was used to perform limiting dilution on 50% of the standard, and each gradient was diluted twice with the negative sample, and then 11 samples were tested.
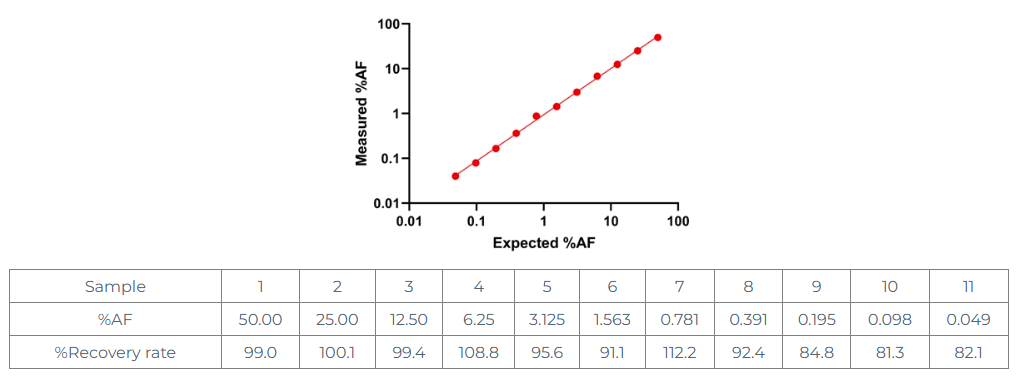
From the above data, it can be seen that when AF is in the range of 0.05%-50%, the corresponding recovery rate is in the range of 80-120%, which is in line with expectations.
Experiment 5:Precision test
Select 20 samples with strong positive rate of 2%, 20 samples with medium positive rate of 1%, and 20 samples with weak positive rate of 0.2% for digital PCR detection.
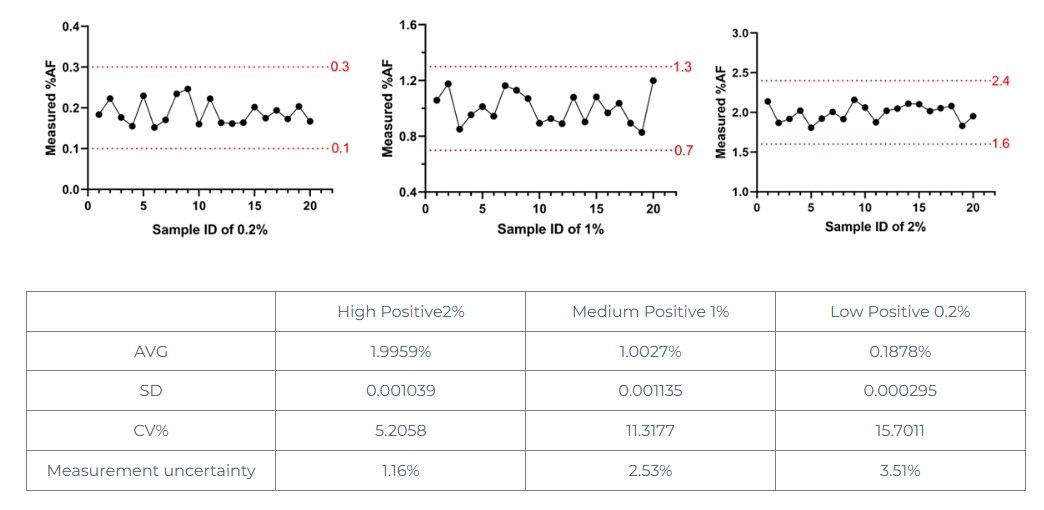
From the above data, it can be seen that under the three detection systems of strong positive 2%, medium positive 1% and weak positive 0.2%, the errors are all within a reasonable range and the precision is good.
Experiment 4:Detection limit test
We can predict the possible detection limit range through theoretical calculation. When the control copy is within 15000 (corresponding to 50ng input), the LOB data is 2, 2/15000=0.0133%, and the predicted detection limit is 0.0200%.
In such a low frequency, the clinical application scenario is suitable for ctDNA. Generally speaking, in clinical practice, the median ctDNA of 10ml whole blood is 20-40ng, and the lowest value is 20ng. The theoretical value of 20ng is 6000 copies, 2/6000=0.0333%, and the predicted detection limit is 0.0500%.
The error range of 0.05% is ±50%, which is 0.025-0.075%, so we choose 3 points and explore LODs of 0.010%, 0.050%, and 0.100%, respectively, requiring 95% detection.
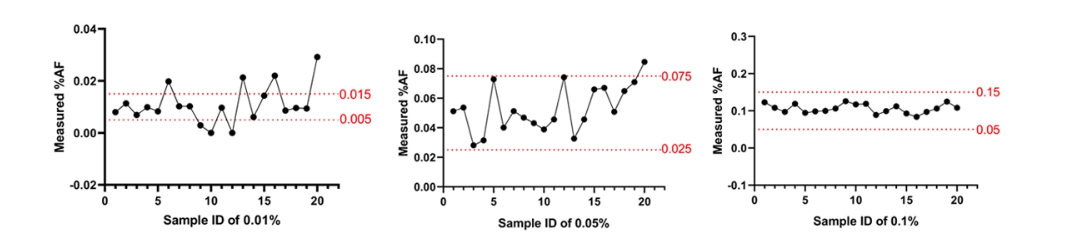
0.010%
Perform digital PCR test on 20 samples with 0.01% to see if 95% detection is achieved within 0.005-0.015%
0.05%
Digital PCR testing was performed on 20 0.05% samples to observe whether 95% detection was achieved within 0.025-0.075%.
0.1%
Digital PCR testing was performed on 20 0.1% samples to observe whether 95% detection was achieved within 0.05-0.15%.
Digital PCR testing was performed on 20 0.05% samples to observe whether 95% detection was achieved within 0.025-0.075%.
0.1%
Digital PCR testing was performed on 20 0.1% samples to observe whether 95% detection was achieved within 0.05-0.15%.
From the above data, it can be seen that 0.05% is selected as the detection limit LOD of the detection kit.
Experiment 3:Linear range test
1.Linear test between input amount and copy number and frequency
First, select the EGFR T790M standard (CBP10402) with a mutation frequency of 50%. This standard is a two-allele gene with CN=2 edited by gene editing. One is mutation and the other is WT. After theoretical value calculation, Sanger sequencing, and Bio-Rad DdPCR kit (Assay ID: dHsaCP2000019), it is confirmed to be 50% frequency.
1.1 Set 10 gradients between 0.1ng and 200ng input amount, and perform digital PCR detection (2 replicates) to consider whether the FAM copy number is linearly related to the input amount and whether the VIC copy number is linearly related to the input amount;
1.2 Set 10 gradients between 0.1ng and 200ng input amount, and perform digital PCR detection (2 replicates) to determine whether the fluctuation of %AF=50±10% is satisfied.
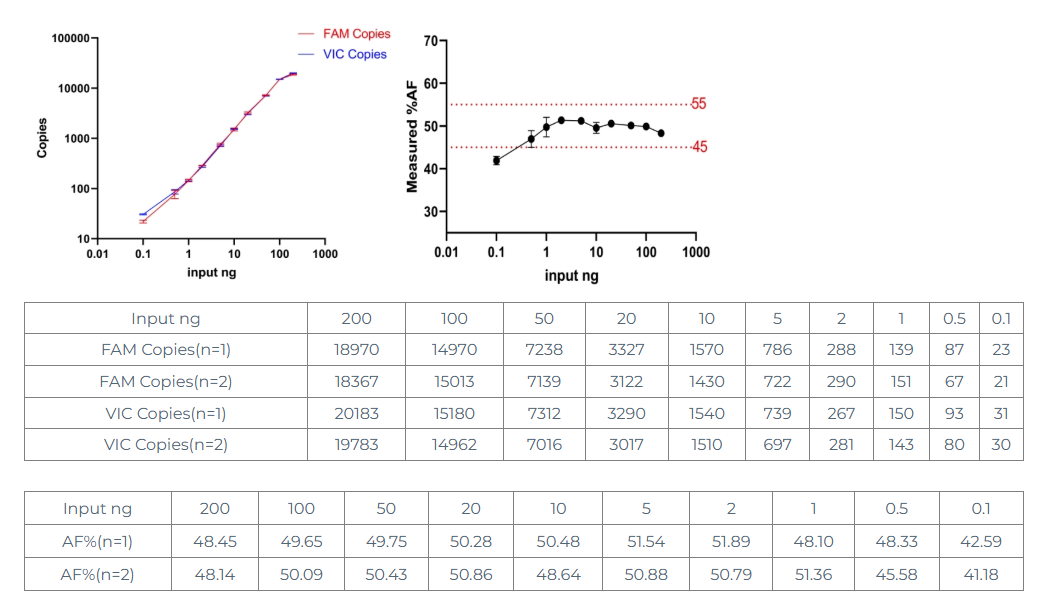
From the above data, we can see that:
①200 ng input is too large, the copy number is not linear, although it meets the range of 45-55%, it is not suitable for single channel copy number determination;
②0.1 ng input is too small, the copy number is not linear, and 42.59% is also lower than 45%, which is not suitable for single channel or dual channel;
Therefore: the range is 0.5-100ng
2.Linear test of theoretical values and detection values at different frequencies of linear dilution
Based on the above conclusions, we selected 10ng as a further exploration of the %AF linear range. First, we used the corresponding negative standard of EGFR T790M, performed limiting dilution on 50% of the standard, set 9 AF gradients between 0.1% and 50%, and performed digital PCR detection (2 replicates)
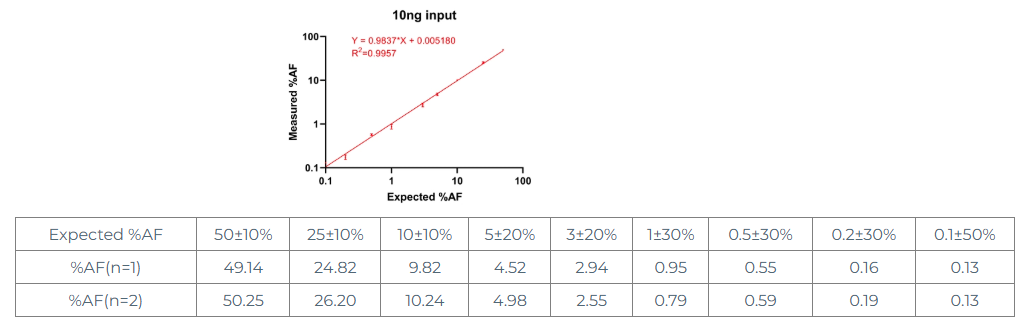
From the above data, we can see that at an input of 10 ng, AF% shows very good linear dilution, with some fluctuations, but all within the error range.
Experiment 2:Accuracy test
Samples to be tested: 10 samples verified as positive by NGS (numbered 1-10); 10 samples verified as negative by NGS (numbered 11-20). Digital PCR was used to test the 20 samples.
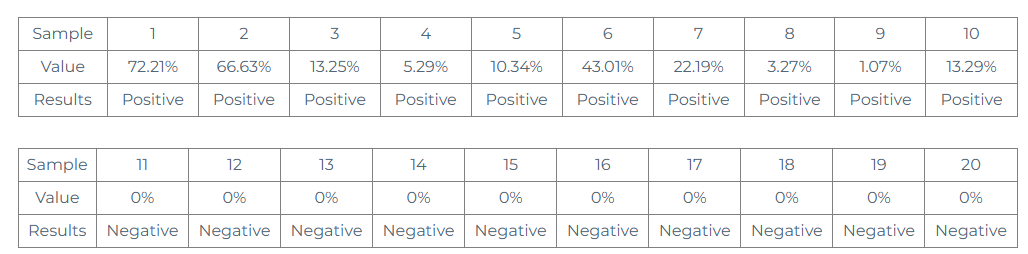
From the above data, we can see that:
① Test 10 NGS positive samples, the test results are all positive, the positive compliance rate is 100%, and the variation frequency is quantitatively detected.
② Test 10 NGS negative samples, the test results are all negative, the negative compliance rate is 100%, and the fixed variation frequency is 0%.
Experiment 1:LOB blank limit test
Samples to be tested: 20 negative samples and 1 NTC. The input amount was set to 10ng and 40ng respectively, and each sample was tested by Bio-Rad digital PCR in duplicate.
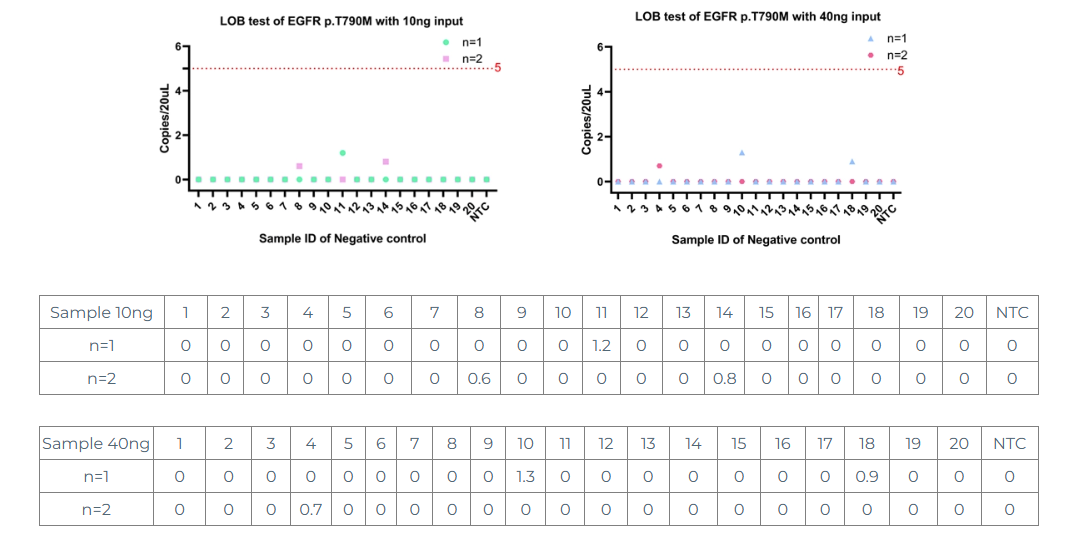
From the above data, it can be seen that whether it is 10ng or 40ng, LOB is controlled within 2 copies (generally the noise copies are within 5), and the noise does not show dose dependence and is a randomly generated noise signal.
Other stable cell lines
If you are interested in ordering, please contact us.
Customer help-line
4008-750-250
sales@reqbio.com
Office address:
3rd Floor, No. 6, Lane 222, Guangdan Road, Pudong New Area, Shanghai, China
We Are Pleased to Announce: Global Commercial Licensing Rights for Jurkat E6.1, CHO-K1, and HEK293 Cell Lines Officially Secured.
Explore